DNA Replication and Repair
A solid foundation in cellular and molecular biology begins with a thorough understanding of DNA replication. Moreover, it is essential to comprehend the mechanisms of DNA repair and the phenomenon of DNA shortening.
Click on the labels to gain in-depth knowledge about each component. To explore further, double-click or click on another component to navigate away. For a comprehensive understanding of other major cellular processes, visit the page on transcription and translation.
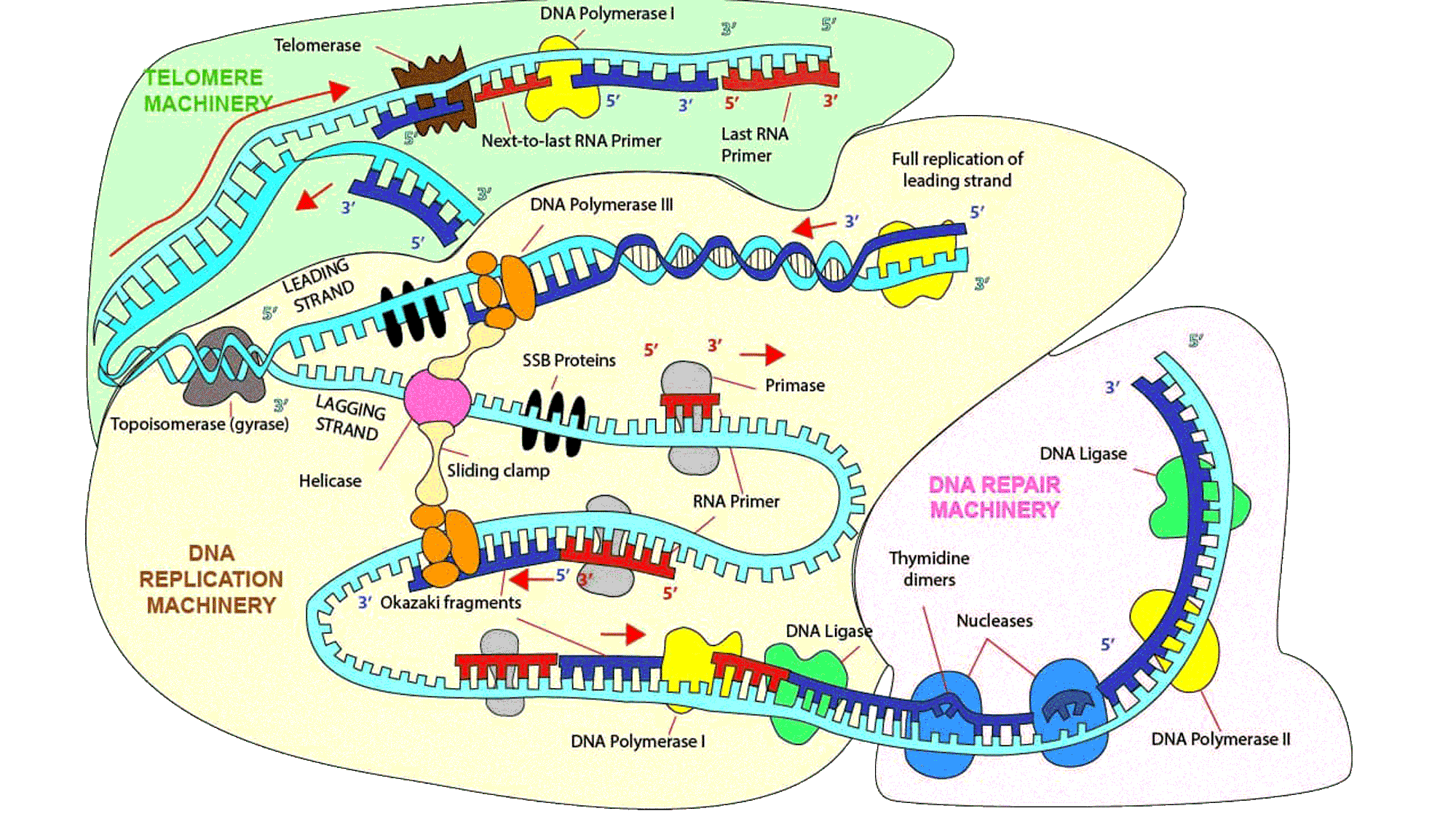
DNA Polymerase III
DNA polymerase III holoenzyme is the major enzyme involved in replication. It also proofreads the daughter DNA via its 3' to 5' exonuclease function and replaces errors through its 5' to 3' synthesis.

Single-strand DNA binding (SSB) proteins
These proteins stabilize the unwound single-stranded DNA and modulates the function of replication, recombination and repair machinery.
DNA Polymerase I
It is a processive enzyme that sequentially catalyzes multiple polymerization reactions while holding the DNA template strand. It performs proofreading of erroneous replicons, synthesis of correct sequences, and removal and replacement of RNA primer with DNA to fill the Okazaki fragment gaps. Ribonuclease H and flap endonuclease I (FEN I) are its eukaryotic counterparts.
RNA Primase
After helicase unwinds parent DNA at the origin, a specialized RNA polymerase called primase forms a short ~12 nucleotide long RNA primer complementary to unwound template strand.
DNA Polymerase II
It acts as a backup enzyme for replication. It has 5' to 3' synthesis activity and 3' to 5' exonuclease activity in prokaryotes.
DNA ligase
It forms a phosphodiester bond between the 3' OH end of an acceptor nucleotide and 5' phosphate end of a donor nucleotide during replication (combining Okazaki fragment nicks), repair (binding after error is resolved) and recombination of DNA (in chromosomes during meiosis).
Okazaki Fragments
DNA polymerases can add nucleotides to the growing daughter strands only in 5' to 3' direction. Copying of lagging template strand must proceed in opposite direction from the replication fork. Each 100-200 nucleotide attaches to an RNA primer and elongates 5' to 3' in discontinuous DNA segments called Okazaki fragments.
Nucleases
These enzymes cleave the phosphodiester bonds between nucleotides. The proofreading domain in DNA polymerase ensures that base-pairing stops when an incorrect base is added, 3' end is transferred to its exonuclease site, 3' end is removed, and the DNA strand is sent back to the synthesis domain to add the correct base. Nucleases can also be independent of the polymerase, like the restriction endonucleases.
Helicase
In order to act as a template, parent DNA duplex is unwound by helicase which starts at the origin of replication. It is usually an AT-rich sequence.
Topoisomerase
Local unwinding of duplex DNA produces torsional stress, which is relieved by topoisomerase I, while the simultaneous unwinding of DNA duplex occurs through helicase.

DNA Repair
There are broadly 3 categories of DNA repair - base excision repair, in which a base is replaced by another base (pre-replication repair), mismatch excision repair, in which mismatched, inserted or deleted bases, maybe a few nucleotides long, are repaired (post-replication repair); and nucleotide excision repair, which fixes chemically modified bases that distort DNA shape locally.



Double stranded DNA breaks are caused by ionizing radiations (X-rays and gamma rays) and a few anticancer drugs. It is severe and can cause cellular and genetic abnormalities. These are repaired using homologous recombination and non-homologous end joining (NHEJ), the latter being error-prone and susceptible to lost of base-pairs. This phenomenon can induce genetic mutations.

DNA Shortening
All known DNA polymerases elongate DNA chains at the 3′ end, and all require an RNA or DNA primer. As the replication fork approaches the end of a linear chromosome, lagging-strand cannot be replicated in its entirety. When the final RNA primer is removed, there is no upstream strand onto which DNA polymerase can build to fill the resulting gap. Without some special mechanism, the daughter DNA strand would be shortened at each cell division.

DNA Errors
DNA damage can occur by (i) spontaneous cleavage of chemical bonds (ii) ultraviolet and ionizing radiation (iii) toxic chemical by-products in the cell.
How does DNA damage occur?
(i) DNA polymerase inserts wrong nucleotide - mutation (ii) copying errors (iii) genetic aberrations during recombination causing repair machinery to fail - tumor , cancer or genetic disorders.

Why is a telomere machinery needed?
During replication, while leading strand synthesis continues till the end of the DNA template, lagging strand is not copied in its entirety. When final RNA primer is removed, there is no upstream strand on which DNA polymerase can build to fill the resulting gap.
Telomerase
An enzyme telomere terminal transferase or telomerase adds telomeric repeats at chromosomal ends. This is determined by telomerase-associated RNA and not the source of telomeric DNA primer. Through its own RNA, a telomerase performs telomere sequence recognition. Once attached, it adds a few oligoribonucleotides to the telomeric DNA via reverse transcription.
